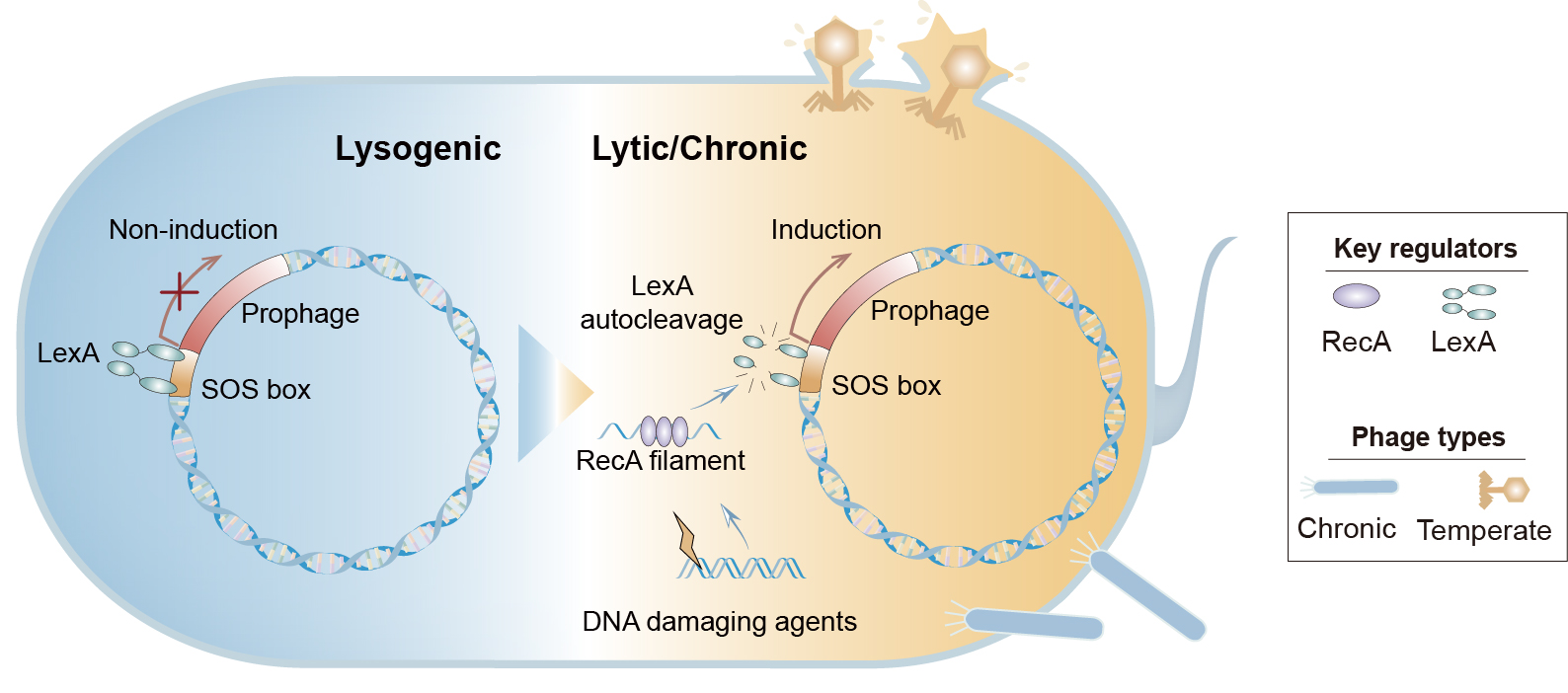
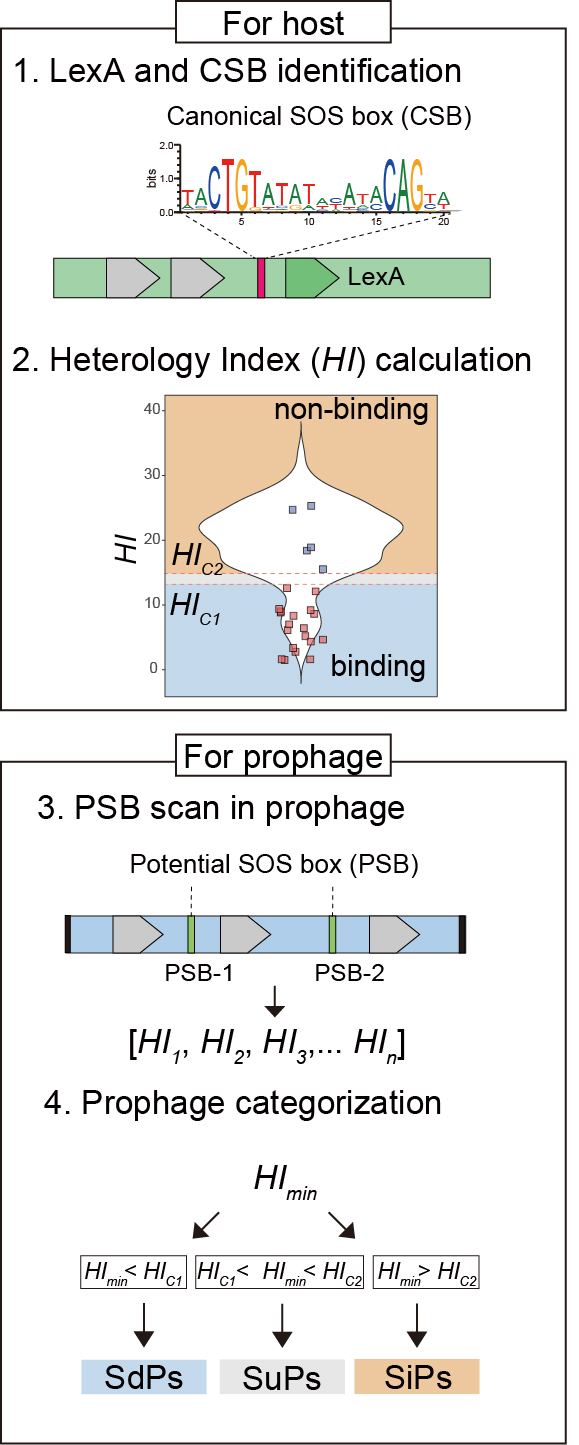
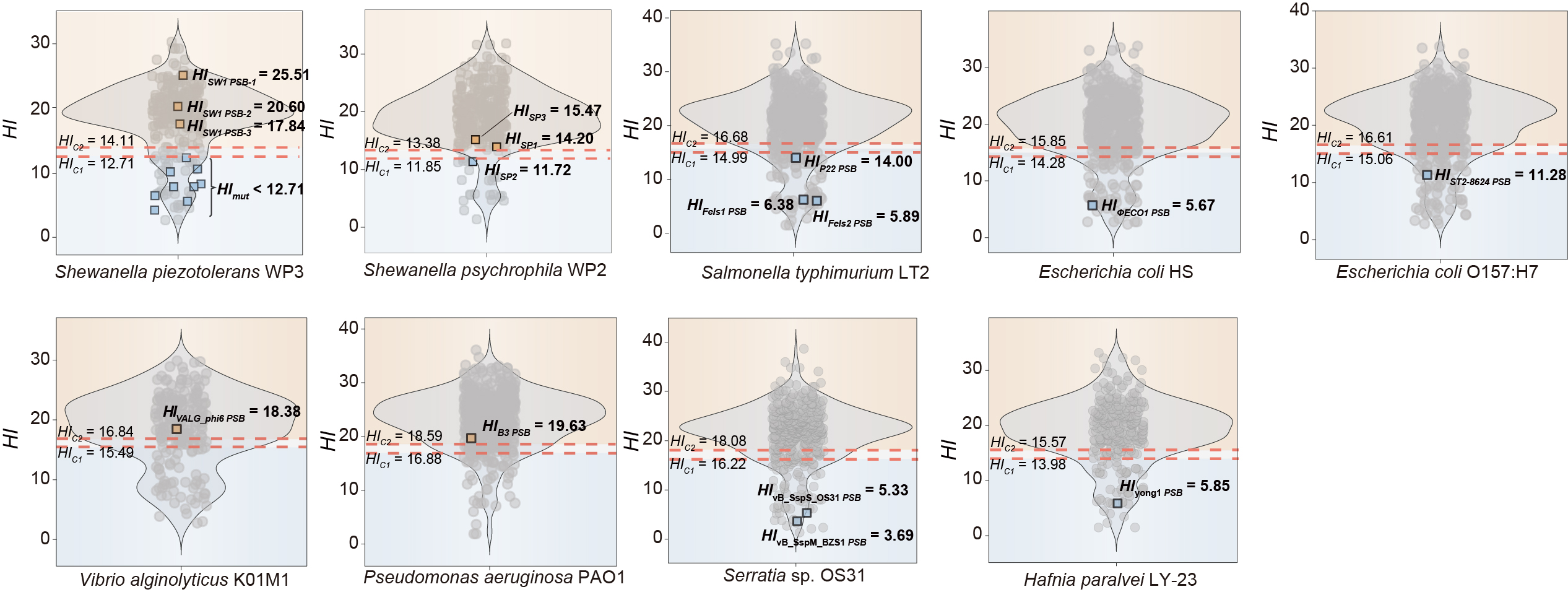
The PSOSP workflow consists of four main steps, leading to a detailed output:
- LexA & Canonical SOS Box (CBS) Identification : Scanning the host genome to identify LexA protein and canonical SOS boxes (CSBs) located upstream of the lexA gene.
- Heterology Index (HI) Calculation: Identifying potential SOS boxes (PSBs) across bacterial genomes, calculating the Heterology Index (HI) for each PSB and establishing classification thresholds (HIc1 and HIc2) via Mean Shift clustering results.
- PSB scan in prophage: Scanning PSBs within prophage promoter regions and determining of the minimum HI (HImin).
- Prophage categoriation : Evaluating the ability of LexA binding to prophage promoter regions by comparing HImin with thresholds